
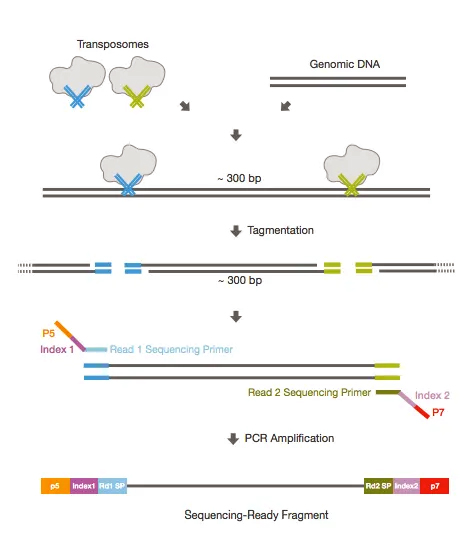
Recently, high-throughput or next generation sequencing (NGS) technologies enabled metagenomic-based identification of viruses by sequencing random fragments of all genomes present in a clinical or environmental sample. However, specific PCR requires prior knowledge of the virus sequence, and a separate assay needs to be designed for each individual virus or virus type. To date, sequence-specific PCR is the most common approach for virus identification and quantification in diagnostic laboratories as it is highly sensitive, rapid, and cost effective. Our protocol supplements existing virus-specific detection strategies providing opportunities to identify atypical and novel viruses commonly not accounted for in routine diagnostic panels. The workflow for virus metagenomic sequencing that we established proved successful in detecting a variety of viruses in different clinical samples. Our sequencing protocol does work not only with plasma but also with other clinical samples such as urine and throat swabs. Our method proved successful in detecting the majority of the included viruses with high read numbers and compared well to other protocols in the field validated against the same reference reagent. We further validated our method by sequencing a multiplexed viral pathogen reagent containing a range of human viruses from different virus families. A protocol including filtration, nuclease digestion, and random amplification of RNA and DNA in separate reactions provided the best results, allowing reliable recovery of viral genomes and a good correlation of the relative number of sequencing reads with the virus input. In order to optimize metagenomic sequencing for application in virus diagnostics, we tested different enrichment and amplification procedures on plasma samples spiked with RNA and DNA viruses. Here, we describe a comprehensive sample preparation protocol for high-throughput metagenomic virus sequencing using random amplification of total nucleic acids from clinical samples. Recently, advances in high-throughput sequencing allow for virus-sequence-independent identification of entire virus populations in clinical samples, yet standardized protocols are needed to allow broad application in clinical diagnostics.
However, as specific PCR only detects pre-defined targets, novel virus strains or viruses not included in routine test panels will be missed. Sequence-specific PCR is the most common approach for virus identification in diagnostic laboratories.


 0 kommentar(er)
0 kommentar(er)
